I recently worked through Udacity’s Data Engineering nanodegree program which consisted of four lessons: Data Modeling (PostgreSQL and Cassandra), Data Warehousing (Redshift), Data Lakes (Spark), and Pipeline Orchestration (Airflow).
In this post, I’ll share some of my notes from the third lesson: Data Lakes. This lesson plan focused on using HDFS with Spark which are both open-source data lake software packages and not specific to a particular cloud platform. However, every cloud platform has their own service for operating the clusters of hardware that run Spark and HDFS. For AWS it’s EMR, and for GCS it’s DataProc. This post will go into greater detail of the AWS services.
What is a Data Lake?
Let’s review the types of data stores that have already been covered in the previous posts. Relational databases are vertically scalable, individual computers for processing queries on data stored on disk memory (one query per CPU). NoSQL databases are horizontally scalable clusters of computers that process queries in a distributed (isolated) fashion. Data Warehouses, like Redshift, are also distributed clusters and horizontally scalable, but process queries in a parallelized fashion across all CPUs in the cluster (remember these are called MPP databases for “massively parallelized”).
Data Lakes are essentially a way of achieving the kind of parallelization that Data Warehouses achieve, but on commodity hardware (and therefore, lower cost), as well as storing and processing unstructured data. Because they operate at lower cost, Data Lakes can be HUGE and store and process vast amounts of data. The name for Data Lakes comes from an analogy between a bottle of water and a lake. Data Warehouses are like bottles of water in that they are clean and neatly packaged and self-contained. Data Lakes on the other hand can be HUGE compared to the water bottle but contain a lot of unwanted garbage.
The core technologies of the Data Lake are the Hadoop Distributed File System (HDFS) and Apache Spark (like everything in IT infrastructure, there are many, many alternatives, but these are two of the main software packages for Data Lakes). The former is for distributed storage and the latter is for distributed processing. The incredible thing about Data Lakes is that they infer “Schema-on-Read” which means the HDFS files can be queried like a normal, running database as the files are being read. For this reason, the typical ETL process is sometimes described as ELT for Data Lakes, because the Transformation of the data happens after it is already Loaded into the HDFS. Spark is also capable of reading directly from compressed gzip files. Note that the order of ELT means the data in the Data Lake is stored “as-is” without any cleaning applied to it, giving rise again to the analogy of a dirty lake. But having access to the original data is quite a boon for Data Scientists wanting to run ad-hoc queries and train machine learning models on Big Data. With a Data Lake, Data Scientists aren’t limited to just the highly preprocessed data in the Data Warehouse.
Because they can get so messy, Data Lakes haven’t completely replaced Data Warehouses, but they do have several crucial advantages. Among these advantages is the fact that HDFS can store unstructured (e.g. images, large text files) and semi-structured (json) data along with structured (tabular) data, while Data Warehouses mostly just store structured data. I also just mentioned how easily machine learning integrates with Spark. Because Spark is an open-source software, developers have integrated some open-source Machine Learning libraries into Spark, causing Spark to become synonymous with “scalable” machine learning applications. Later in this post, I’ll show an example of one such library, spark-nlp.
With these remarks in mind, lets revisit the original ETL pipeline that I’ve been referencing and see how it might look as an ELT pipeline:
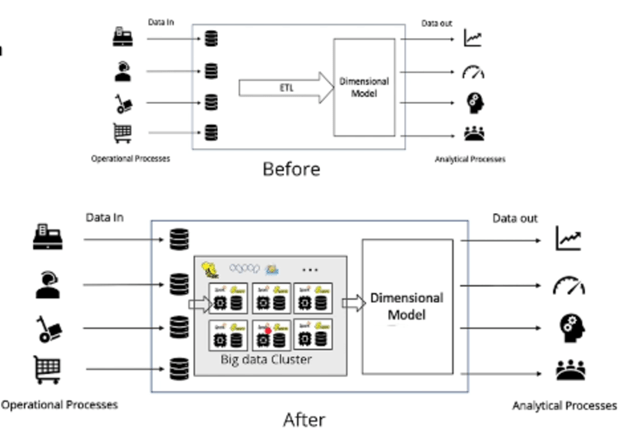
The illustration above shows the replacement of ETL from being done on one machine to being performed in a distributed fashion. The Data Warehouse and the Data Cubes can remain the same with the addition of the Data Lake to this pipeline, but new capabilities have been added. The ETL, as well as any ad-hoc queries and Machine Learning, can be run on a distributed Spark Cluster which, since it’s running on commodity hardware, has near infinite scalability.
Data Lakes on AWS
Like everything in the cloud, there are multiple ways to deploy a Spark Data Lake in the cloud:
- AWS EMR with HDFS and Spark (traditional Data Lake)
- AWS EMR with S3 and Spark (24/7 access to storage and highly scalable storage)
- AWS Athena with S3, AWS Glue, and AWS Lambda (cloud-native/serverless)
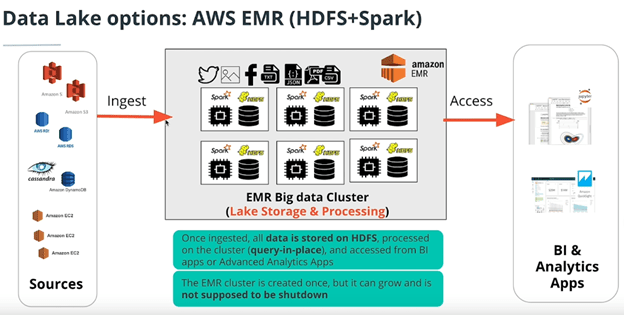
One problem of having your storage on your cluster is that if your cluster is off, you don’t have access to your database. Compare that to S3 where you have access all the time.
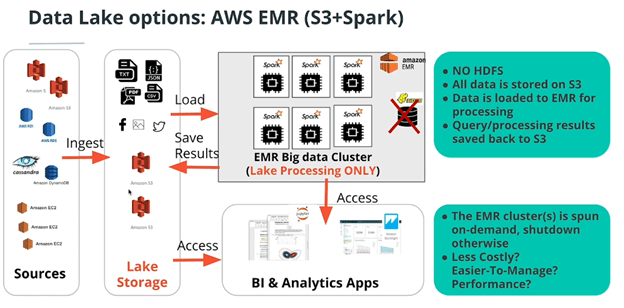
The main benefit of this is you don’t have to keep your cluster running all day to have access to the dataset. You just turn it on for the transformation and then turn it off (like a step function). The trade-off for this cost saving is the increase in latency for having to load the entire dataset from S3 into the Spark cluster for processing. For production quality pipelines, the S3 bucket must be in the same AZ and region as the EMR cluster, and even then, this might not be advisable.
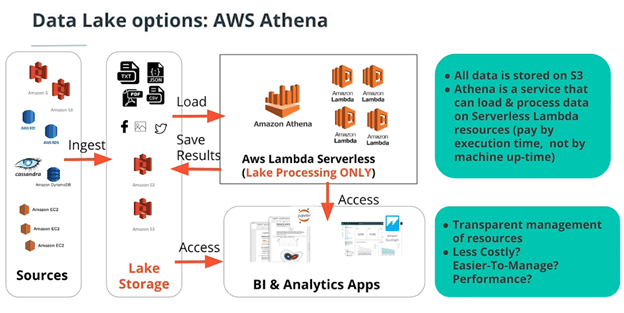
Lambda is a “function as a service”. Instead of provisioning EC2 instances, you just pay for the time to run your code. What AWS EMR is to EC2 instances, Amazon Athena is to Lambda functions. It loads the data, automatically allocates the right amount of compute power, and then executes it on Lambda. It’s the completely serverless way of running an ETL pipeline.
One way to run Athena is to use AWS Glue which is AWS’s schema-on-read data store. It infers an entire database schema from the file structure of the dataset in S3. Inferring data types and relations and everything (where each table is a subfolder with a collection of csvs in it that all have the same schema). In this cloud native example of an ETL pipeline on AWS, data flows from S3 → Glue → Athena → Lambda → Redshift → BI dashboard.
Notice that these three options represent different benefits and trade-offs in terms of both latency and cost. If the processing needs of a pipeline are small but storage requirements are large, then the serverless option might be best. If computation need are high and the Spark cluster will have a high availability, then using HDFS with the Spark cluster is probably the best option.
Natural Language Processing with Spark-NLP
One of the many nice features of AWS EMR is the ability to seamlessly run notebooks on your Spark Cluster. Within the EMR page, the left side bar has an option for “notebooks”. By creating a new notebook on this page, you will be able to develop and test your Spark code directly on your EMR cluster. Here is an example of Spark code that uses the Machine Learning package spark-nlp to perform part-of-speech filtering on a dataset of Reddit articles.
# Perform a word count for all the nouns (common and proper) in the dataset of Reddit article Titles
#start spark session
from pyspark.sql import SparkSession
spark = SparkSession.builder.config("spark.jars.packages","JohnSnowLabs:spark-nlp:1.8.2").getOrCreate()
#load all the files in the reddit dataset
file_path = "./data/reddit/*.json"
df = spark.read.json(file_path)
dfAuthorTitle = df.select("data.title", "data.author")
#Use spark-nlp to do pos tagging
from com.johnsnowlabs.nlp.pretrained.pipeline.en import BasicPipeline as bp
dfAnnotated = bp.annotate(dfAuthorTitle, "title")
#each word in the title column has now been annotaed by spark-nlp's basic pipeline to include the token the lemmatized text, and the part of speech id.
# Use the part of speech annotations to filter for only Nouns in the title
#explose the tokenized sentences into a single column
from pyspark.sql import functions as F
dfPos = dfAnnotated.select(F.explode("pos").alias("pos"))
#filter for only nouns (plural or singular)
filter = "pos.result = 'NNP' OR pos.result = 'NNPS'"
dfNNP = dfPos.where(filter)
# Exapand the single column into multiple columns with select expression
dfWordTag = dfNNP.selectExpr("pos.metadata['word'] as word", "pos/result as tag")
#perform the final word count
from pyspark.sql.functions import desc
dfWordCount.groupBy("word").count().orderBy(desc("count")).show()
Additional spark-nlp resources:
- https://towardsdatascience.com/introduction-to-spark-nlp-foundations-and-basic-components-part-i-c83b7629ed59
- https://medium.com/spark-nlp/spark-nlp-101-document-assembler-500018f5f6b5
- https://medium.com/spark-nlp/spacy-or-spark-nlp-a-benchmarking-comparison-23202f12a65c
- https://towardsdatascience.com/natural-language-processing-with-pyspark-and-spark-nlp-b5b29f8faba
Criticisms of the Data Lake:
The ability to bring Machine Learning into the world of Big Data is a huge win for Data Lakes, but they are not without their downsides and trade-offs. As I mentioned before, they can get rather messy and unorganized (sometimes referred to as data garbage dumps). The “all are welcome” motto to take data “as-is” can lead to people throwing a lot of trash in the Data Lake. This makes Data Governance rather difficult. If the file structures and naming conventions are unorganized, how could anyone hope to determine who has access to what data.
Additionally, the relative disorganization of HDFS compared to Data Warehouse schemas, can lead to significantly more shuffling of data. This shuffling is done over network connections within the same region and AZ, but still, network connection are roughly 20x slower than an ordinary SSD read.
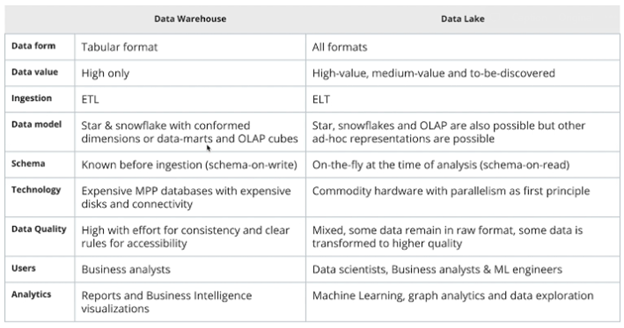
The Udacity course summarizes the comparison between Data Warehouses and Data Lakes in the following table. Ultimately, like all data stores, each has its own strengths and weaknesses, and should be deployed in its proper time and place.
That’s it for now. Next up, scheduling data pipelines with Apache Airflow.